Introduction
Spatial transcriptomics technologies provide high-resolution spatial maps of transcript locations within tissue. A central challenge in these datasets is cell segmentation, which assigns transcripts to individual cells accurately, particularly when image data is noisy, missing, or ambiguous.
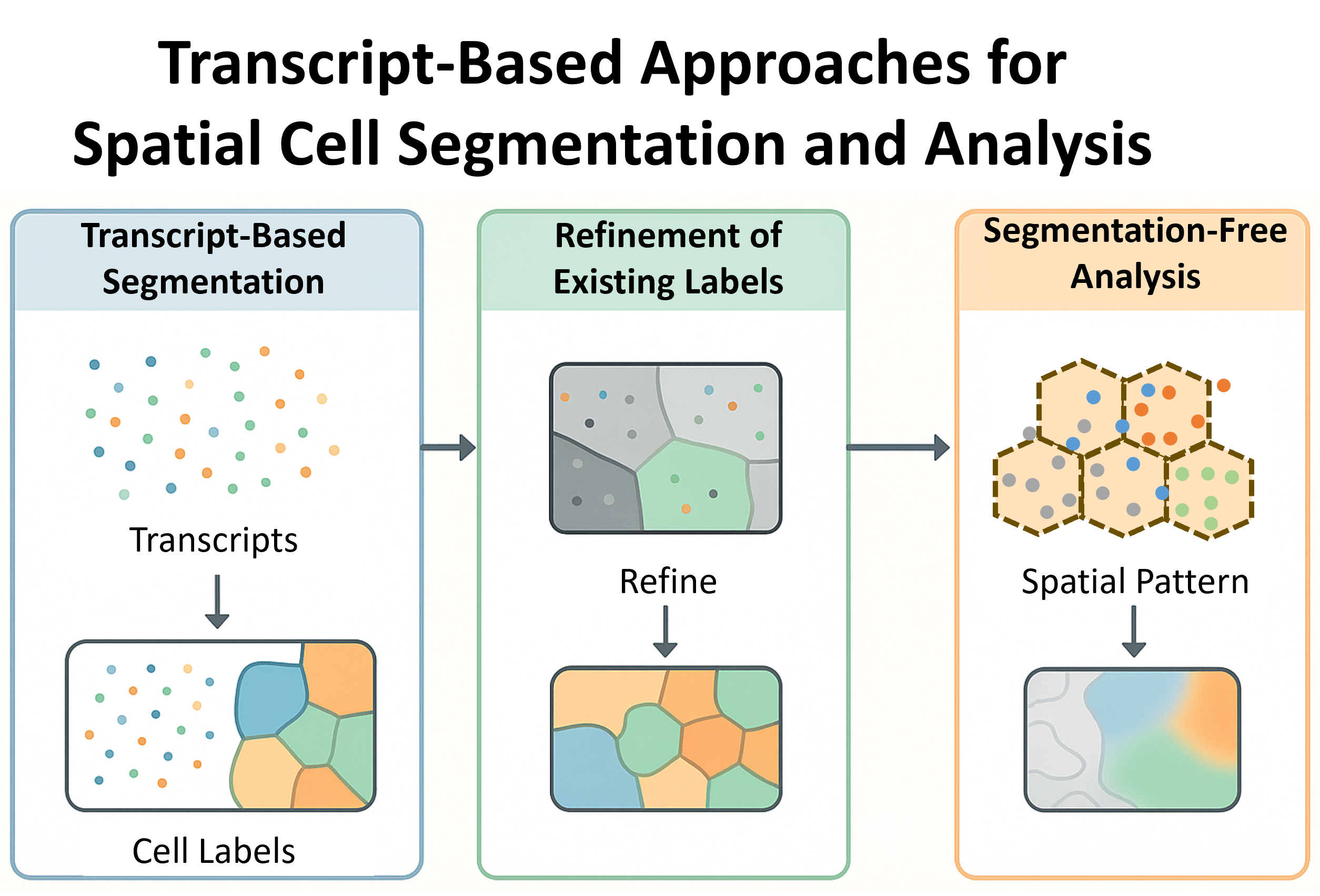
To address this, researchers have developed transcript-based analysis techniques that either:
- Section 2.1 Segment cells from transcript locations (e.g., Baysor, ProSeg),
- Section 2.2 Refine existing image-based segmentations using transcript patterns (e.g., FastReseg),
- Section 2.3 Avoid segmentation entirely by analyzing the spatial organization of transcripts directly (e.g., FICTURE).
This post provides a practical walk-through of each technique with examples on using it with CosMx® data, whose standard data format are described here.
Like other items in our CosMx Analysis Scratch Space, the usual caveats and license applies.
Choosing the Right Transcript-Based Approach
Spatial transcriptomics platforms differ in resolution, density, and image quality, and so do the challenges in analyzing them. Before diving into individual tools, it’s helpful to understand the three major transcript-informed approaches to working with spatial data: segmentation, refinement, and segmentation-free analysis. Each method type fits different scenarios and solves a unique class of problems.
Transcript-Based Segmentation
These methods directly infer cell boundaries by clustering or modeling the spatial distribution of transcripts, often using gene identity as additional signal.
Use when:
- You don’t have reliable cell images (e.g., missing or low-quality DAPI/membrane stains)
- You want to define cells purely from mRNA localization
- You need a de novo segmentation pipeline without preprocessing
Limitations:
- May oversegment sparse cells or misplace boundaries without priors
- Transcript noise can bias clustering in low-resolution assays and introduce circularity in analysis pipeline
Best for: Datasets where transcript positions are abundant and dense.
Segmentation Refinement
These tools enhance an existing segmentation mask, correcting common segmentation errors using transcript-level context only when there is sufficient evidence.
Use when:
- You already ran an image-based segmentation (e.g., Cellpose, watershed)
- You notice cells with minor contamination from neighboring cells
- You want to keep image-based alignment but improve transcript association
Limitations:
- Relies on the quality of the initial mask — can’t fix everything
- Adds an extra pipeline step, but is lightweight
Best for: Any pipeline combining tissue images with transcript-based validation, especially if accurate cell boundaries affect downstream quantification.
Segmentation-Free Analysis
Instead of forcing transcripts into discrete cells, these approaches model gene expression directly in space, uncovering continuous spatial features, patterns, and regions.
Use when:
- You want to avoid cell segmentation biases
- Your tissue has ambiguous or poorly defined boundaries
- You aim to study gradients, niches, or expression domains more than individual cells
Limitations:
- No per-cell outputs (e.g., no cell-by-gene matrices)
- Some tools are exploratory and require interpretation beyond standard stats
Best for: Ultra-dense data like Seq-Scope or CosMx where transcript resolution enables high-fidelity spatial patterning without segmentation artifacts.
References
Jones, D. C., A. E. Elz, A. Hadadianpour, et al. 2025.
“Cell Simulation as Cell Segmentation.” Nature Methods.
https://doi.org/10.1038/s41592-025-02697-0.
Petukhov, Vladimir, Rui J. Xu, Ruslan A. Soldatov, Pietro Cadinu, Konstantin Khodosevich, Jeffrey R. Moffitt, and Peter V. Kharchenko. 2021.
“Cell Segmentation in Imaging-Based Spatial Transcriptomics.” Nature Biotechnology.
https://doi.org/10.1038/s41587-021-01044-w.
Si, Yichen, ChangHee Lee, Yongha Hwang, Jeong H. Yun, Weiqiu Cheng, Chun-Seok Cho, Miguel Quiros, et al. 2024.
“FICTURE: Scalable Segmentation-Free Analysis of Submicron-Resolution Spatial Transcriptomics.” Nature Methods 21 (10): 1843–54.
https://doi.org/10.1038/s41592-024-02415-2.
Wu, Lidan, Joseph M. Beechem, and Patrick Danaher. 2025.
“Using Transcripts to Refine Image Based Cell Segmentation with FastReseg.” Scientific Reports 15 (1): 30508.
https://doi.org/10.1038/s41598-025-08733-5.