1 Introducion
I am consistently impressed by the remarkable cellular resolution, sensitivity, and comprehensive spatial profiling capabilities that CosMx® SMI technology offers. Indeed, I often analyze datasets that consist of 10s of slides and millions of cells – numbers that are steadily growing with time.
At several points in my analysis workflow I need to subset the data. For example, I may want to examine a specific niche or ask what the molecular composition of each kidney glomerulus looks like. While some of these subsets can be achieved by computational means, there are some scenarios where I isolate specific regions or specific cells by directly selecting them on the tissue. I refer to this as selecting Regions of Interest (ROIs).
In this post, I will show you how to define ROIs using the napari-cosmx plugin. In a future post, we’ll show a similar approach using R.
1.1 Preliminaries
This is the fourth installment of our napari series. Readers should be familiar with the napari-cosmx plugin and the basics. I’ll also make use of the python interpreter and users may benefit from reviewing the post on stitching before reading further.
2 Selecting Groups of Cells
There are two ways to interact with napari and the napari-cosmx plugin. The first is via the GUI (see napari-cosmx plugin and the basics for a refresher). If you’re working with the GUI, drag and drop your CosMx SMI napari folder into napari’s main window to view the tissue. Once opened, you can access the ipython terminal by clicking on the >_ icon which is located on the bottom left of the application (see yellow arrow in the previous post). The main object that we’ll work with is named gem (for Gemini).
For users more comfortable with directly launching napari and napari-cosmx using the command line, this second method is for you. Activate your virtual environment containing the napari-cosmx plugin. I’m testing this with Python 3.9, pyenv, napari 0.5.4, PyQt5 5.15.11, and napari_CosMx-0.4.17.1. After activation, launch ipython and then instantiate your gem object.
# in bash
cd to_project_directory
pyenv activate your_virtual_environment_name
ipython# in ipython/python
from importlib.metadata import metadata
import numpy
import pandas as pd
import os
from napari_cosmx.gemini import Gemini
import pickle
from os import listdir
import imageio
data_dir = '/path/to/napari/data/files'
gem = Gemini(data_dir)In my example dataset, I have the metadata already loaded.
gem.metadata cell_ID cell_type initial_cell_type leiden
UID
10101 c_1_100_1 B cell 1 B cell 1 0
10200 c_1_100_100 CD4 T cell CD4 T cell 2
1000100 c_1_100_1000 B cell 1 B cell 1 0
1002101 c_1_100_1001 CD4 T cell CD4 T cell 2
1004104 c_1_100_1002 B cell 1 B cell 1 0
... ... ... ... ...
350859 c_1_395_592 CD4 T cell CD4 T cell 2
20457783 c_1_254_4523 B cell 1 B cell 1 0
3261907 c_1_271_1806 B cell 2 B cell 2 1
4670103 c_1_182_2161 plasma cell plasma cell 12
25664444 c_1_88_5066 plasma cell plasma cell 12
If you are working directly in ipython, you can open up the right hand widget using the show_widget method.
Let’s color cells by cell_type. I prefer the terminal for this task as I have finer control of the individual colors for each cell type.
my_colors = {
"B cell 1": "#AA0DFE",
"B cell 2": "#3283FE",
"CD4 T cell": "#85660D",
"B cell 3": "#782AB6",
"fibroblast reticular cell 1": "#565656",
"fibroblast reticular cell 2": "#1C8356",
"macrophage": "#16FF32",
"pericyte": "#F7E1A0",
"GC B cell": "#E2E2E2",
"CD8 T cell": "#1CBE4F",
"interferon-stimulated cell": "#C4451C",
"APC 1":"#DEA0FD",
"plasma cell":"#FE00FA",
"NK cell":"#325A9B",
"nonspecific":"#FEAF16",
"epithelial cell":"#F8A19F",
"APC 2":"#90AD1C",
"dendritic cells":"#F6222E"
}
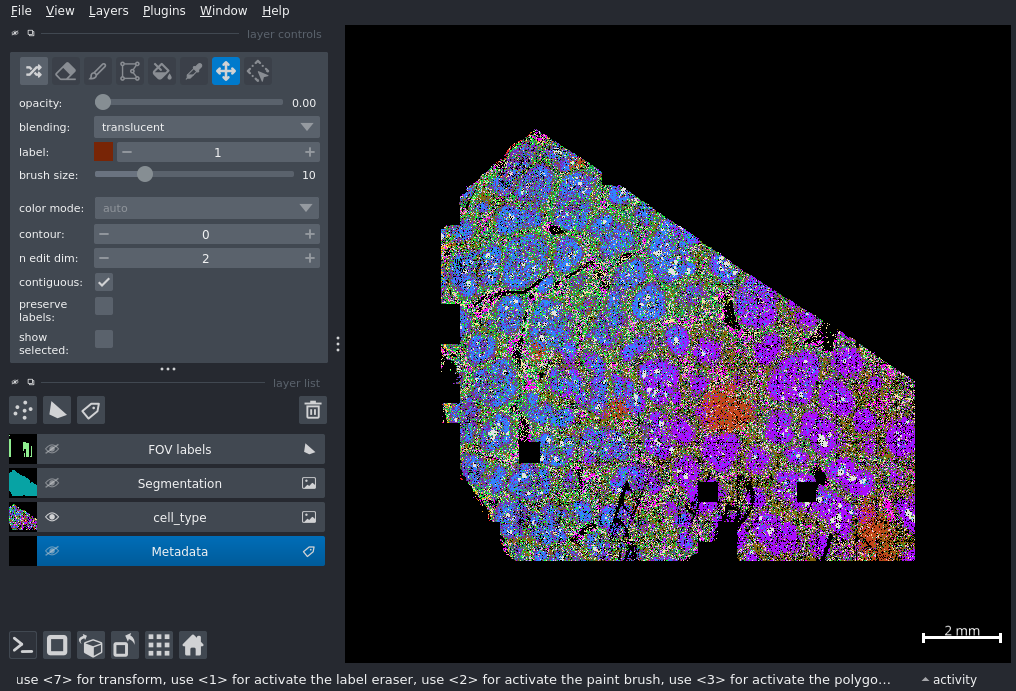
gem.viewer.layers['Segmentation'].visible = False
gem.viewer.layers['FOV labels'].visible = False
gem.color_cells("cell_type", color=my_colors)
# Optional step
fig_path = "./fig-cell-types.png"
with imageio.get_writer(fig_path, dpi=(250, 250)) as writer:
screenshot = gem.viewer.screenshot(canvas_only=True)
writer.append_data(screenshot)At this point, your image should look like Figure 1.

Let’s create a single ROI around one of the nodules. Here are the steps:
- Zoom and pan to desired region
- Create a Shapes layer by clicking the filled white polygon on the left side panel.
- Select face color and edge color. Tip: to show no face color, set the alpha to 0.
- Click on the shape type and draw polygon
- Optional: save ROI to disk
Steps 1-4 can be seen in the video below.
We can rename that layer by double clicking it or with the command below.
gem.viewer.layers['Shapes'].name = 'ROI001'We can also save this ROI as a pickle file so we can later come back to it when we open the sample again.
To save:
gem.save_layers(['ROI001'], "path/to/output/data")And if we want to load that file later:
gem.load_layers(['ROI001'], "path/to/output/data")2.1 More efficient ROI creation
There are many nodules in this sample. While the process of creating an ROI is simple, this manual process can take time. It can be more efficient to look at computational approaches like defining these nodules based on shared cell type composition (i.e., niche). But if we still want fine control of the ROIs, we can speed up this process by creating a few helper functions that do many of these steps for us.
In the code below, we are creating a keyboard shortcut that will create a new shapes layer simply by pressing n on your keyboard. The new layer will be named ROI001 if it’s the first one; otherwise, it will add an integer to the ROI name (ROI002, ROI003…). After creating a new layer, pressing p will activate the polygon tool allowing you to select the vertices of a given polygon. Double-clicking the last vertex will “close” the polygon.
I tend to keep the shapes layer inactive whenever possible. This limits the number of accidental vertex additions to a shape.
Here’s the code. Be sure to add the shortcut definitions after creating the gem object. You can change the color and edge width to any valid value. Here I’ll make the border yellow with a width of 100.
def find_next_roi(my_list):
"""Finds the next ROI code in a list of ROI strings.
Args:
my_list: A list of strings in the format "ROIxxx" where xxx are digits.
Returns:
The next ROI code in the sequence, or "ROI001" if the list is empty
or the next sequential code if no gap is found.
"""
if not my_list: # Handle empty list
return "ROI001"
# Extract numeric parts and convert to integers
numbers = [int(item[3:]) for item in my_list]
# Sort the numbers in ascending order
numbers.sort()
# Find the first gap in the sequence
for i in range(len(numbers) - 1):
if numbers[i + 1] - numbers[i] > 1:
next_number = numbers[i] + 1
return f"ROI{next_number:03d}"
# If no gap is found, return the next sequential number
next_number = numbers[-1] + 1 # Get the last number and increment
return f"ROI{next_number:03d}"
# This keybinding makes it easier to create a new shape layer for ROIs
@gem.viewer.bind_key('n', overwrite=True)
def new_roi_layer(viewer):
# Search layers for "ROI" pattern
layers = viewer.layers
names = [x.name for x in layers]
names_filtered = [item for item in names if "ROI" in item]
new_roi_name = find_next_roi(names_filtered)
print(f"Creating {new_roi_name} layer.")
viewer.add_shapes(
shape_type='polygon',
scale = [gem.mm_per_px, gem.mm_per_px],
name=new_roi_name,
face_color='transparent',
edge_color='yellow',
edge_width=100
)
In the video below, I am quickly creating a new shapes layer using the keyboard combination n + p.
And now that I have four ROIs, I can save them like this:
N=4
roi_list = ['ROI'+str(i).zfill(3) for i in range(1, N+1)]
gem.save_layers(roi_list, "path/to/output/data")2.2 Subsetting Based on ROIs
What we would like to do now is to identify which of the cells are in each of these polygons. There’s a method available in napari-cosmx to do just that.
Using the roi_list list we created above, we can generate a Boolean column in the metdata for all of the ROIs.
Depending on the size of the sample, the size of the ROI(s), and your hardware, this process can be computationally intense.
new_metadata = gem.layers_to_metadata(roi_list)
new_metadata cell_ID cell_type initial_cell_type ... ROI002 ROI003 ROI004
UID ...
10101 c_1_100_1 B cell 1 B cell 1 ... False False False
10200 c_1_100_100 CD4 T cell CD4 T cell ... False False False
1000100 c_1_100_1000 B cell 1 B cell 1 ... False False False
1002101 c_1_100_1001 CD4 T cell CD4 T cell ... False False False
1004104 c_1_100_1002 B cell 1 B cell 1 ... False False False
... ... ... ... ... ... ... ...
350859 c_1_395_592 CD4 T cell CD4 T cell ... False False False
20457783 c_1_254_4523 B cell 1 B cell 1 ... False False False
3261907 c_1_271_1806 B cell 2 B cell 2 ... False False False
4670103 c_1_182_2161 plasma cell plasma cell ... False False False
25664444 c_1_88_5066 plasma cell plasma cell ... False False FalseRecall that you can save your new metadata file to disk so that it can be used later with the color cells widget. (e.g., new_metadata.to_csv("_new_metadata.csv", index=False)).
ROIs can be hierarchical. For example, one can create concentric rings around a focal region. So a given cell can be True for multiple ROIs.
If you prefer to have these extra columns converted into a single column with the name of the ROI a given cell is found in, you can use this function.
def generate_roi_summary_column(df):
"""
Generates a 'ROI' column based on all ROI columns in the dataframe and removes the individual ROI columns.
Args:
df: The pandas dataframe containing ROI columns.
Returns:
The updated dataframe with the 'ROI' summary column and individual ROI columns removed.
"""
# Get all columns that start with 'ROI'
roi_cols = [col for col in df.columns if col.startswith('ROI')]
# Filter dataframe to only include ROI columns
roi_df = df[roi_cols]
# Create a new column 'ROI' and initialize it with None
df['ROI'] = None
# Iterate over each row
for index, row in roi_df.iterrows():
# Check if all values in the row are False
if not any(row):
continue # Keep 'ROI' as None
# If there's at least one True, find the first column name where it's True
for col in roi_cols:
if row[col]:
df.loc[index, 'ROI'] = col
break
# Drop the ROI columns
df.drop(columns=roi_cols, inplace=True)
return dfHere’s a tabulation of the number of cells found in each ROI in this example.
meta_summary = generate_roi_summary_column(new_metadata)
meta_summary.ROI.value_counts()ROI
ROI001 13503
ROI003 8536
ROI002 7686
ROI004 5721
Name: count, dtype: int64As you can see in the code above, in the case you have hierarchial ROIs, a given cell that is present in multiple ROIs will be assigned the first ROI in the list.
3 Selecting Individual Cells
Sometimes I want to focus on individual cells instead of regions. A typical case for me is when I want to select “anchor” cells for use with InSituType. And while the above polygon-based ROI selection approach does work on single cells, it’s a little more effort than just clicking a cell.
In the code below, I zoomed into an arbitrary position, replaced filled polygons with cell type contours, and turned on a few IF channels.
gem.viewer.layers['cell_type'].visible = False
gem.color_cells("cell_type", color=my_colors, contour = 1)
gem.viewer.window.set_geometry(0, 49, 1440, 773)
gem.viewer.camera.center = (0.0, 2.2981841637346654, 12.99023480800836)
gem.viewer.camera.zoom = 7477.634103126872
gem.add_channel('CD68', colormap = 'red')
cd68 = gem.viewer.layers['CD68']
cd68.contrast_limits = [1560.7958115183246, 5072.586387434555]
gem.add_channel('PanCK', colormap = 'green') # PanCK
PanCK = gem.viewer.layers['PanCK']
PanCK.contrast_limits = [2331.937172774869, 9234.471204188483]
gem.add_channel('Membrane', colormap = 'cyan') # membrane
mem = gem.viewer.layers['Membrane']
mem.contrast_limits = [3684.502617801047, 18176.879581151832]In the video below, I selected a few cells using the Points layer.
Here are those cell IDs.
df = gem.layers_to_metadata(['Points'])
df_filtered = df.loc[df['Points'] == True].cell_ID
df_filteredUID
13697552 c_1_151_3701
14115200 c_1_151_3757
14554376 c_1_151_3815
15437192 c_1_151_3929
15547400 c_1_151_3943
16000151 c_1_151_4000
16329832 c_1_151_4041
19053376 c_1_151_4365
19105792 c_1_151_4371
19132027 c_1_151_4374
Name: cell_ID, dtype: object
4 Conclusion
This blog post provides a step-by-step guide on defining Regions of Interest (ROIs) in CosMx SMI data using the napari-cosmx plugin. It demonstrated how to manually select ROIs with polygons, streamline the process with keyboard shortcuts for efficiency, and subset data based on these selections. I also illustrated how to select individual cells using a Points layer in napari, offering a precise method for identifying cells of interest for further analysis. The code used here has not gone through the typical, vigorous testing process. Please reach out and file a Github issue if you find any bugs.