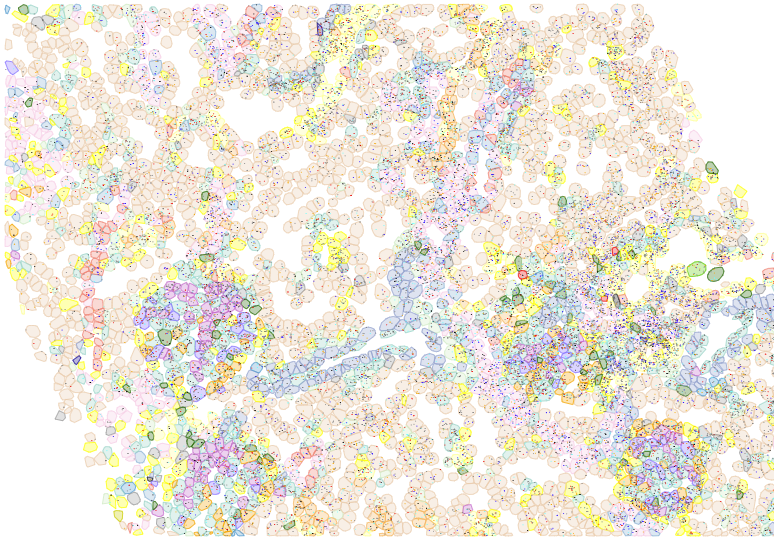
Deriving cell polygons for plotting
A complete CosMx dataset will contain polygonal boundaries for each cell for use in plotting. In practice, especially with earlier datasets or with datasets passed between collaborators, this data can be missing. We’ve created a toolkit for deriving these polygons from cells’ transcript locations. (Note: we are not performing cell segmentation here, just drawing boundaries around transcripts already assigned to cells.)
You can find the package here.
Plotting cells as polygons looks better in zoomed-in views, and it allows for plotting of individual transcripts as in the below: